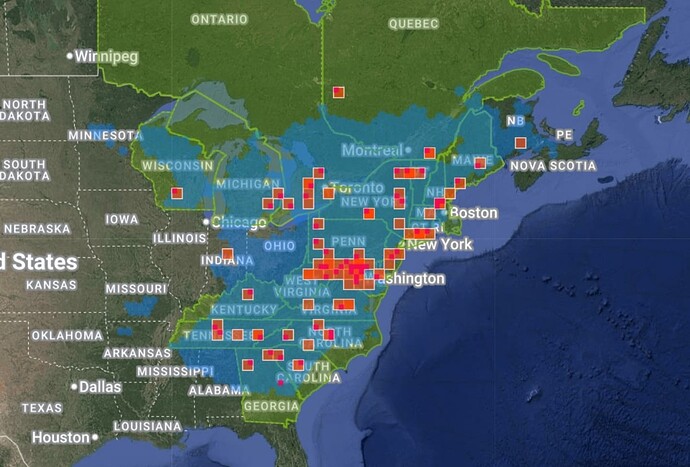
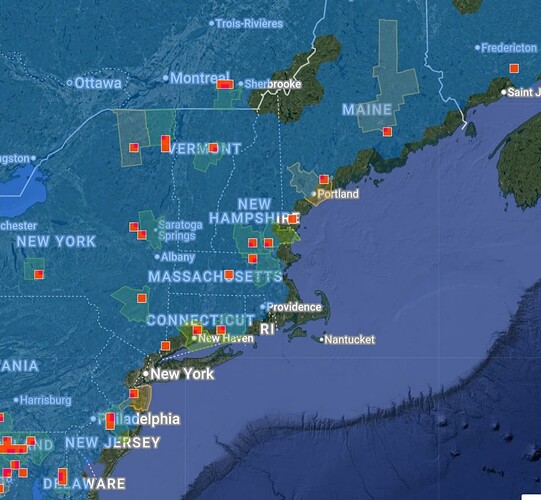
For me, my frustrations with the CV definitely picked up around when the summer flood of observations started coming in (maybe around late May/early June?)
To pick on a specific example that is currently the bane of my existence, let’s take Agelenopsis potteri. Over in eastern Europe and in some parts of Canada, that species is at least possible to ID via typical user-submitted photos, which means there’s a lot of species-level observations for the CV algorithm to get trained on.
But in much of the US, there are extreme lookalikes that are effectively indistinguishable or require an incredibly discerning eye to separate, making species IDs of genus Agelenopsis a much taller task in the USA. As a result, many of those other species in the genus have nowhere near enough photos to be put into the CV as competitors for suggestions.* We will be sidestepping the whole “but wait they actually can’t even be separated by external photos anyway” concern for now.
*This is another aside that is not directly relevant to Tony’s question, but I would like to get it off my chest. Many of the observations that do exist for those other species are of preserved specimens, which have a distinctly different look that I certainly wouldn’t trust the CV to try to extrapolate out to “the appearance of live spiders that haven’t been dunked in alcohol,” so even if there happened to be enough collected specimen observations to get into the algorithm, it wouldn’t help for all of the live photos that people are taking.
I have noticed a consistent uptick in observations with Agelenopsis potteri CV suggestions this summer that are completely unjustified for the level of detail visible in the observation (presumably because the CV algorithm looks at all the European and Canadian observations and goes “Oh, this must always be easily identifiable to species by image matching”), so I’ve had to kick more and more of them back to genus all summer long.