Its new web site is theoretically designed for ongoing updates, but to my knowledge that has not been put into practice yet. There are still a few volumes awaiting publication, then maybe they will have more resources to start updating older treatments.
https://forum.inaturalist.org/t/nature-inspired-memes/27300
and there are more …
(Acacia - means thorn - for the thorn trees of Africa - which are now Senegalia and Vachellia - while Acacia means Australian wattle with not a thorn in sight)
Those two statements aren’t necessarily at odds. It’s entirely possible that two species (good “biological” species–those that refuse to mate with one another in natural settings) can’t be distinguished phenotypically but can be distinguished genetically. In which case, the published description includes the genetic characters. Mother nature doesn’t care whether we humans can distinguish them easily. They’re not competing paradigms–merely the reality we live in. And that reality can be very, very challenging for us humans. But we do the best we can to describe and document that reality (beyond what we can see with our eyes).
I’ve seen people include nucleotide sequence as part of the diagnosis (Gaga) but I didn’t think a genetics-only diagnosis would be valid under the botanical Code. There is a tentative move to amend that for fungi that have no reproductive structures but I am not sure that will pass.
I remember someone doing a genetics-only classification of a genus of parasitic wasps and getting roundly criticized in literature for it for the reason Charlie gave earlier: you just wind up creating a genetic taxonomy in parallel with the morphological taxonomy and it becomes impossible to integrate the two.
I didn’t think “can only be identified by sequencing” applied except for bacteria, where degree of DNA-DNA hybridization is a criterion for separation (due to the relative lack of morphology and asexual reproduction).
That standard hasn’t been used for species for a long time at least in vascular plants. There are many, many ‘species’ that freely intergrade.
From a pragmatic sense, many genii are huge, so putting something to genus is nearly useless. For instance, Carex. There are about 100 species here. Identifying Carex sp. is not very valuable. Versus Polytrichum, a fern with two ‘species’ that are nearly identical. I always identify those to genus already. I don’t think they are valid species, but given there are only two in the genus, it doesn’t matter too much. If the splitters split genuses as well, and made a new genus for each cryptic species set, it would solve some issues but create a bunch of others… and change itself causes problems too. Not just the ‘i dont like change’ kind, the ‘database hopefully corrupted’ kind or ‘i can’t use hsitoric data any more’.
Good question, i am not sure.
ooh, i want a dark beer. wait then we can debate which beers really count as dark.
That’s great, but why dismiss others who have concerns of that level? You realize just cuz you don’t have a problem doesn’t mean no one else does either. Maybe i overstate too much but i promise you it isn’t only me who struggles with these issues.
Would having them at subspecies level cause you problems? If not, why not do that?
I guess when i say ID on sight i am not ruling out bringing something back home and using a dissecting scope or something. So maybe i haven’t been communicating well. And yes for tiny non vascular plants, diagnostic features are bound to be tiny. Some of these splits go way beyond that though.
Yeah, i just don’t think the Asarium one affected me directly, i’m sure i’d disagree with it. But yeah my point isn’t that field botany shouldn’t be hard. it’s that it’s already so hard with understaffing, multiple crises, etc, why make it any harder just for the sake of splitting to species instead of subspecies. Like, we have X number of hours a day and X number of brain cells, so splitting Caulophyllum probably has literally led to some wetland being bulldozed, some population going extinct, etc, because we had to spend our time on fussing over databases and returning to sites to find flowers instead of doing more important conservation work. Sure, the taxonomists can and should do this detailed work, but with hypersplitting they literally force it on everyone else (or else stop people from doing ecology). I just can’t understand anyone seeing that as a worthwhile tradeoff when there’s literally a solution that still allows them to document fine-scale differences in populations through subspecies and variety. Like, no one has ever been able to explain to me the downside other than literally splitting species to manipulate endangered status or else they just like saying they ‘discovered’ a species, You’re here in good faith Choess. What is the argument against using subspecies here? Cuz it confuses me.
This can be difficult, the short answer is i am usually in wetlands and pallida is very very rarely in wetlands here, i’ve seen it maybe once in a high elevation enriched seep. I do think the leaves look different, but maybe it isn’t consistent. Also i generally don’t expect a functionally full species list except Mid June-Mid Sept and usually i can figure out capensis in that time. I got in an argument about it on iNat once with someone who bumped me back to genus for small Impatiens capensis in a seep where pallida wouldn’t be. Himalayan balsam is still barely in Vermont, but as it spreads that will make it harder for sure :( but ‘please don’t do this to other things’ is valid too. Like with Caulophyllum and Arisaema, those often don’t have flowers during my main survey window so they are harder. And while the Arisaema subspecies does have a different environmental correlate it doesn’t seem important enough to break all the old data over it.
i wonder why my experience is so different. Maybe i’m in a bubble, people in Vermont don’t tend to like change. I definitely think it’s made iNat fall apart, elsewhere i just don’t do the splits, at least for now.
I see most CoC applied to subspecies too though. So it doesn’t answer that whole question. I think there are all kinds of cryptic populations that mess with CoC. I literally deleted red maple from my own special CoC calculation because i think it indicates literally nothing, it’s in the best and the worst condition wetlands alike. I us ethe metric a lot but it’s certainly imperfect. I think i’d be more invested in going the other way and finding ways to merge difficult to distinguish species that have similar CoC into meta-units or something.
nor does she care if something is at species vs subspecies level :D
I also wonder if evolutionary convergence can happen at a genetic level. As in, can genetics truly definitively define past evolution? Surely genes change at different rates and maybe certain genes are maintained by epigenetics or else certain sequences co-evolve in less related species because they are useful. I find the premise of defining species solely on genetics possibly problematic.
There was a project that was collecting leaves from I. pallida and I. capensis to study them and see if there was any detectable difference - I wonder if that yielded any results?
Impatiens capensis is in North America only?
That taxonomist was confused - capensis for Southwestern Cape. In Africa.
I think that having Symphyotrichum firmum included in S puniceum does cause problems (granted these are admin problems, but problems nonetheless). Right now S puniceum s.l. is listed by army corps as a OBL plant, but in my region this is just wrong, it’s more like a FAC plant because S firmum is a FAC/FACU plant and counting by area more common. I manage a well drained to excessively well drained former corn field next to a gravel pit where it is bone dry with 10+% cover of an OBL plant. It in its own little way breaks the system. If we did everything at leaf taxa level (CoC, Wetlands indicator status, conservation status…) I would have almost no complaints, but the decision by taxonomic end users (agencies and techs) seems to be more on the side of treating species like monolithic entities. If we are going to treat them as one thing then they need to describe just one thing not a bunch of ecologically distinct vars.
I think part of the problem at this point is new species are described, but there is a lag time of sometimes decades before we get useful regional keys for these taxa, and that lag can be a real struggle. And sometimes after all that time the keys still suck, and you still can’t easily know the species. I’m sure there are plenty of plants that you just know out of bloom, that would be impossible to determine from existing keys. For me learning what these splits actually describe has made each one more palatable for me. The Caulophyllum split has been on my priority list to understand this spring, and I think my problem is that I only seem to have the segregate giganteum where I botanize most of the time. I’m hoping that as I learn the groups trends start to appear that make them appear more different even late in the growing season, but at this point with the keys I have to rely on a few limited characters from their flowers.
I don’t think that the problem with wetland destruction, habitat loss, species extinction, field botanists being overworked, and not enough time in one life has much anything to do with botanical workers spliting. I see that as the product of a culture that prioritizes convenience, comfort, growth, and cost over what is really important: survival, the world we live in, and a sense of human well being. The average person doesn’t seem to care much about the natural world and does not interface with taxonomy at all. Changing that, as Sisyphean a task as that is, seems to me far more productive than creating a more streamlined system to make plant taxonomy more stable.
Yup it was a mistake where the describer thought it was collected in South Africa. Asclepias syriaca is another geographic mistake that jumps to mind.
Not all heroes wear capes, but all continents have one
FYI: The plural of genus is genera.
No way S. puniceum should be OBL, that doesn’t make sense here either. I’d say Fac-Wet here, it prefers wetland but does fine in mesic uplands.
well then it’s an impossible choice because either you make it so that you can’t even do the army corps plots most of the time, or else you can’t get the right precision for your results. I mean the solution is really within ACOE not taxonomy for that one, but even i know that’s pretty hopeless.
let me know if you figure out caulophyllum. to me it’s such a great example of a subspecies or variety because they intergrade so widely. Yes that’s odd when they don’t necessarily overlap that much with blooming, but i still observe it often. There must be overlap.
Oh i agree, but something that really throws piles of wrenches on top of field botanists given all of this other stuff definitely causes its own harm. But i am not saying splitters are putting wal-marts in wetlands or something. They care about conservation too hey just have a fundamentally flawed and easily fixable approach that needs to be tweaked so we can all get what we need.
Oh capes are a dime a dozen.
Ours is - the
Fairest Cape in the whole circumference of the earth.
The Cape.
But since this is iNat
Cape Floral Region
This x1000. Everyone going rogue and using a different name for the same taxa is way more confusing and worse than just leaving a group of species at genus level while the folks closest to that group take the time to make an informed hypothesis about how to organize that group in light of new information.
As we come up with new tools for determining the relationships among taxa sometimes that sort of explodes some groups we had organized using other methods, and that’s ok, it’s all a work in progress with the hope that each new technology and taxonomic change gives us a little more information to make sense of the world around us. Sometimes folks are very resistant to change and reject new names simply because it’s different rather than being excited that we have gathered more taxonomic information. I get that, I get attached to species names and sometimes my knee jerk reaction is to be annoyed that now it has a new name I have to remember when I prefer the old name, especially for species I am fond of. But I often come to realize that the new change makes a lot of sense, and with a little time those fond feelings for a species associate with the new name too. It helps to frame it as something gained rather than something lost.
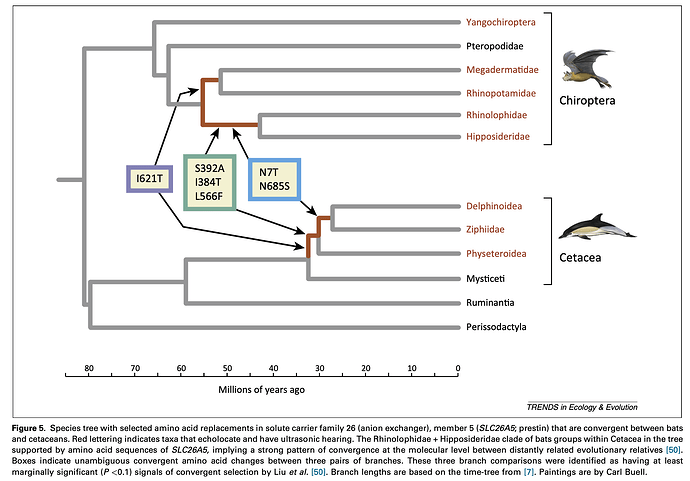
Molecular convergence has been demonstrated in some groups, such as in bats and whales. The figure below is from McGowen et al. (2014).
McGowen, M. R., J. Gatesy, and D. E. Wildman. 2014. Molecular evolution tracks macroevolutionary transitions in Cetacea. Trends in Ecology and Evolution 29:336-346.
It’s important to remember that convergence in the genome only effects certain genes. The whales and bats converge in genes related to echolocation, but otherwise they are different. Similarly, whales, fish, penguins, and seals converge in shape because they all move fast through water, but you have no trouble telling them apart. Well, a little trouble with the fish/whale difference but still, with a close look you find them different. Any one gene can give you a false phylogeny. This is why biologists studying relationships are moving toward whole-genome sequencing even though it’s more expensive and results are a bear to analyze.
i guess that brings us back full circle to AI with all that data to look at…
Evidently dictionary.com hasn’t refreshed their taxanomic meta data. Go figure!
Genus is an English word and thus may be pluralized according to the rules of English. The fact that it is a Latin loanword does not give it an intrinsically special status forcing it to be pluralized according to the rules of Latin; we use English pluralizations of loanwords from virtually all other languages, regardless of how they would have been pluralized in the language they are borrowed from. That said, because most of the people pluralizing the word ‘genus’ in a professional setting are also Latin nerds on some level, they may recognize you as an outsider if you do not use the Latin pluralization.